snomedizer: R Interface to the SNOMED CT Terminology Server REST API
08 July 2022
Source:vignettes/snomedizer.Rmd
snomedizer.RmdOverview
What is SNOMED CT?
SNOMED CT is the international standard ontology of clinical terms. It can be thought of as an encyclopedia on all things relevant to medicine, for instance: anatomical structures, diagnoses, medications, tests, surgical procedures, pathogens such as SARS-CoV-2 or Candida auris, or even clinical findings, such as a high temperature.
SNOMED CT is not just a list of terms. It is an ontology made up of concepts which are fully defined in relation to each other. This allows complex inferences illustrated throughout the present vignette. You can learn more about ontologies in this introduction workshop [1].
SNOMED CT is currently used in more than 80 countries. It is present in over 70% of clinical systems commercialised in Europe and North America [2,3], and it is mandated across the English National Health Service for:
- records of symptoms, diagnoses, procedures, medications, observations or allergies [4]
- electronic prescribing and pathology laboratory systems [5,6].
The best ways to learn about SNOMED CT are:
- the SNOMED International Browser
- the SNOMED CT for Data Analysts free online learning course
- the official SNOMED CT Starter Guide, particularly chapters 5. Logical Model and 6. Concept Model.
What is snomedizer?
snomedizer is an R package to interrogate the SNOMED CT terminology based on Snowstorm, the official SNOMED CT Terminology Server.
Snowstorm is the back-end terminology service for both the SNOMED International Browser and the SNOMED International Authoring Platform.
snomedizer allows you to perform the same operations as the SNOMED International Browser directly from your R console.
Introduction to SNOMED CT
Browsing the ontology
SNOMED CT contains rich knowledge about medical concepts.
Try for yourself with the quick exercise below:
- Go to the SNOMED CT International Browser and accept licence terms.
- In the ‘Search’ tab, type in
40600002. - Click on ‘Pneumococcal bronchitis’ in the result list.
- In the right panel (‘Concept Details’), click the ‘Details’ tab.
- Look for the relationships: the ontology defines that this concept is a bronchitis, and that the pathological process is infectious (non-infective bronchites exist). It also specifies that the pathogen is Streptococcus pneumoniae.
- Now click on the ‘Diagram’ tab. The same information is displayed graphically. Purple boxes refer to
116680003 | Is a (attribute) |relationships, while yellow boxes notate other attributes of the concept. - In the ‘Refsets’ tab, you will see that this concept is mapped to ICD-10 code J20.2.
Performing advanced queries (ECL)
SNOMED CT has a query language named ‘ECL’ (Expression Constraint Language). Using ECL, users can perform logical searches and learn new facts from any SNOMED CT concept using a range of operators listed in this ECL quick reference table.
For instance, once can search for all the different subtypes of pneumonia. For this, we need to query the descendants of concept 233604007 | Pneumonia (disorder) |.
The corresponding ECL query is <233604007 | Pneumonia (disorder) |, where < is the ECL operator selecting descendants of a concept.
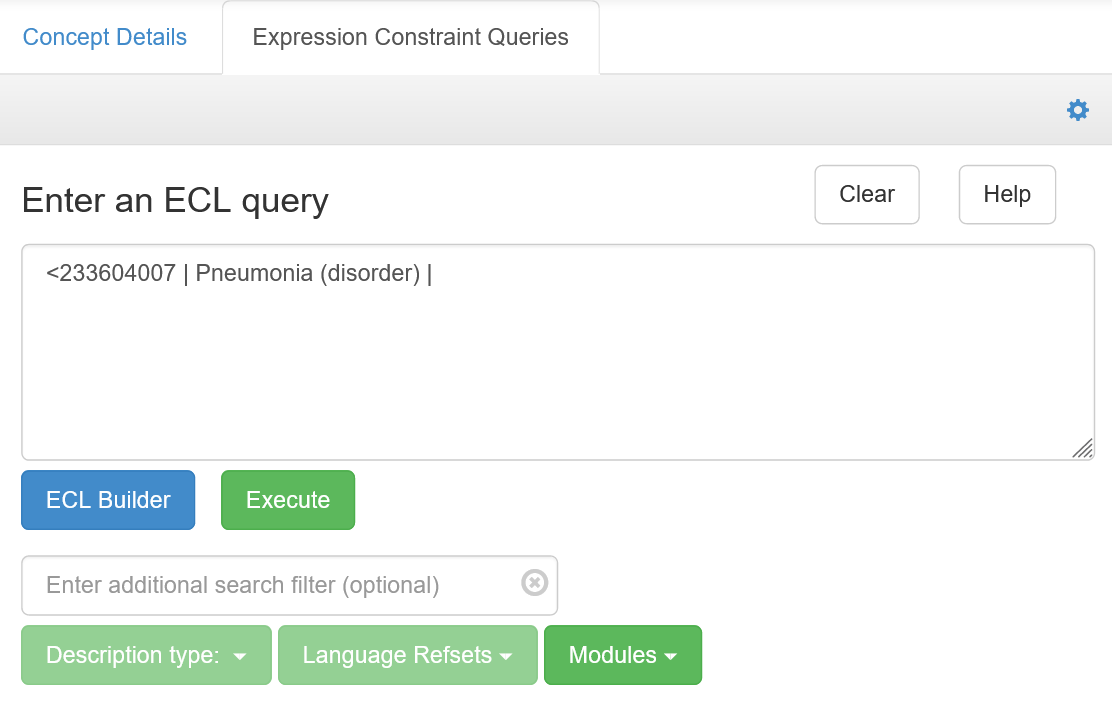
Trying ECL in the web browser
- Navigate to the SNOMED International Browser
- Accept the licence conditions
- Click on the ‘Expression Constraint Queries’ tab in the right panel
- Paste ECL queries from this vignette into the ‘Expression…’ box as shown below
- Press ‘Execute’.
Examples
First, load and set up snomedizer.
## The following SNOMED CT Terminology Server has been selected:
## https://snowstorm.ihtsdotools.org/snowstorm/snomed-ct
## This server may be used for reference purposes only.
## It MUST NOT be used in production. Please refer to ?snomedizer for details.
# Connect to the SNOMED International endpoint
# (see licence conditions in `vignette("snomedizer")`).
snomedizer_options_set(
endpoint = "https://snowstorm.ihtsdotools.org/snowstorm/snomed-ct",
branch = "MAIN/2021-07-31"
)1. Search for relevant urine specimens
Urine tests are commonly performed in hospitals, for instance when looking for bacteria (microbial cultures). Let’s assume we access a laboratory database in which all bacterial cultures are stored with SNOMED CT codes for the specimen type.
There are many types of urine samples, eg: samples of morning urine, mid-stream urine, etc. Some are not optimal samples: for example, urine catheter samples are often contaminated and may give poor information.
Let’s try and
- fetch all the codes corresponding to urine specimens: they are descendants of
122575003 | Urine specimen (specimen) | - while excluding the ones from urinary catheters (
122565001 | Urinary catheter specimen (specimen) |).
First, we need to query descendant concepts. There are two ways to accomplish this in snomedizer:
- by using
concept_descendants() - by running an ECL query in the
concept_find(ecl = "...")function. For this, you will want to use the<<operator (known asdescendantOrSelfOf) using ECL.
All the expressions below are equivalent.
urine_specimens <- concept_descendants(conceptIds = "122575003",
include_self = TRUE)
urine_specimens <- concept_find(ecl = "<<122575003")
urine_specimens <- concept_find(ecl = "<<122575003 | Urine specimen (specimen) |")
glimpse(urine_specimens[, c("conceptId", "pt.term")])## Rows: 40
## Columns: 2
## $ conceptId <chr> "16221491000119104", "16221371000119107", "16221251000119108…
## $ pt.term <chr> "Voided urine specimen", "Urine specimen obtained from pedia…We obtain 40 concepts. But these include some samples obtained from catheters:
## pt.term
## 1 Urine specimen obtained via straight catheter
## 2 Urine specimen obtained via suprapubic indwelling urinary catheter
## 3 Mid-stream urine specimen obtained by single catheterization of bladder
## 4 Urine specimen obtained by single catheterization of bladder
## 5 Urine specimen obtained via indwelling urinary catheter
## 6 Urinary catheter specimenTo exclude those, we make use of the MINUS operator in ECL:
urine_specimens <- concept_find(
ecl = "
<<122575003 | Urine specimen (specimen) | MINUS
( <<122565001 | Urinary catheter specimen (specimen) | OR
<<447589008 | Urine specimen obtained by single catheterization of bladder (specimen) | )
")
glimpse(urine_specimens[, c("conceptId", "pt.term")])## Rows: 34
## Columns: 2
## $ conceptId <chr> "16221491000119104", "16221371000119107", "734443003", "7334…
## $ pt.term <chr> "Voided urine specimen", "Urine specimen obtained from pedia…This now gives us the set of 34 target concepts.
Note: For guidance on ECL operators such as MINUS or OR, see the ECL quick reference table.
2. Find the dose of a medical product
Let’s assume you have electronic prescription records referenced to SNOMED CT medical products. We come across a prescription for SNOMED CT code 374646004, and want to extract the drug type, dose and unit.
Note
Medical product definitions vary considerably across SNOMED CT editions.
In this example, we will use the United States Edition.
med_product <- concept_find(conceptIds = "374646004",
branch="MAIN/SNOMEDCT-US/2021-03-01")
med_product %>%
select(conceptId, fsn.term, pt.term) %>%
glimpse()## Rows: 1
## Columns: 3
## $ conceptId <chr> "374646004"
## $ fsn.term <chr> "Product containing precisely amoxicillin 500 milligram/1 ea…
## $ pt.term <chr> "Amoxicillin 500 mg oral tablet"This prescription is for Amoxicillin 500 mg oral tablets.
We want to extract attributes, which are SNOMED CT relationships giving characteristics of a concept. Attributes can be queries using the . (dot) operator.
First, let’s look at the drug type, which is expressed by attribute 762949000 | Has precise active ingredient (attribute) |.
concept_find(ecl="374646004.(<<762949000 | Has precise active ingredient (attribute) |) ",
branch="MAIN/SNOMEDCT-US") %>%
select(conceptId, fsn.term)## conceptId fsn.term
## 1 372687004 Amoxicillin (substance)We could query other attributes sequentially (or chain them with OR operators in a single ECL expression). But here, the simplest way is to extract all attributes. We do this by remembering that attributes are merely SNOMED CT concepts descending from 762705008 | Concept model object attribute (attribute) |.
med_substance <- concept_find(
ecl="374646004.(<<762705008 | Concept model object attribute (attribute) |) ",
branch="MAIN/SNOMEDCT-US/2021-03-01")
med_substance$fsn.term## [1] "Antibacterial therapeutic role (role)"
## [2] "Tablet (unit of presentation)"
## [3] "500 (qualifier value)"
## [4] "Conventional release oral tablet (dose form)"
## [5] "Amoxicillin (substance)"
## [6] "milligram (qualifier value)"
## [7] "1 (qualifier value)"However, this approach only provides the value of the attribute, not the attribute name.
A more complex, but effective approach involves a special REST endpoint in Snowstorm: GET /{branch}/relationships. We can issue a request to this endpoint directly using the low-level function api_relationships(). Like all low-level api_operations, it is necessary to (1) explicitly request only active concepts, and (2) flatten the output into a data frame. We shall also exclude 116680003 | Is a | attributes (which express inheritance from parent concepts) as they are not relevant in this instance.
drug_attributes <- api_relationships(
source = "374646004",
active = TRUE,
branch="MAIN/SNOMEDCT-US/2021-03-01"
) %>%
snomedizer::result_flatten()
drug_attributes %>%
filter(type.pt.term != "Is a") %>%
select(type.fsn.term, target.pt.term) %>%
arrange(type.fsn.term)## type.fsn.term target.pt.term
## 1 Count of base of active ingredient (attribute) 1
## 2 Has basis of strength substance (attribute) Amoxicillin
## 3 Has manufactured dose form (attribute) Oral tablet
## 4 Has precise active ingredient (attribute) Amoxicillin
## 5 Has presentation strength denominator unit (attribute) Tablet
## 6 Has presentation strength denominator value (attribute) 1
## 7 Has presentation strength numerator unit (attribute) mg
## [ reached 'max' / getOption("max.print") -- omitted 3 rows ]3. Find all diseases caused by a type of bacterium
Let’s extract all infections that can be caused by bacteria belonging to 106544002 | Family Enterobacteriaceae (organism) |.
To do this, we use the : operator, known as ‘refine’ operator.
enterobac_infections <- concept_find(
ecl="<<40733004 | Infectious disease (disorder) | :
246075003 |Causative agent| = <<106544002",
limit = 5000
)
enterobac_infections %>%
select(pt.term)## pt.term
## 1 Bronchopneumonia due to Klebsiella pneumoniae
## 2 Bronchopneumonia due to Escherichia coli
## 3 Salmonella pyelonephritis
## 4 Urinary tract infection caused by Klebsiella
## 5 Infection due to Shiga toxin producing Escherichia coli
## 6 Infection due to Escherichia coli O157
## 7 Septic shock co-occurrent with acute organ dysfunction due to Serratia
## 8 Infection of intestine caused by Salmonella group A
## 9 Infection caused by Citrobacter
## 10 Typhoid peritonitis
## 11 Invasive non-typhoidal salmonellosis
## 12 Sepsis caused by Klebsiella pneumoniae
## 13 Infection caused by Klebsiella
## 14 Meningitis caused by Enterobacter
## 15 Infection caused by Enterobacter
## [ reached 'max' / getOption("max.print") -- omitted 106 rows ]Bonus exercise
Now, extract all body structures that can be infected by the family Enterobacteriaceae.
Tip: you will need the reversed refine operator : R.
Solution
<<123037004 | Body structure (body structure) | : R 363698007 |Finding site| =
(<<40733004 | Infectious disease (disorder) | :
246075003 |Causative agent| = <<106544002)
4. Find synonyms of a concept
Some concepts may have many different names.
Let’s extract synonyms of ‘candidiasis’:
concept_descriptions(conceptIds = "78048006") %>%
.[["78048006"]] %>%
filter(type == "SYNONYM") %>%
select(term)## term
## 1 Candidosis, NOS
## 2 Moniliasis
## 3 Monilia infection
## 4 Thrush
## 5 Moniliasis, NOS
## 6 Infection by Candida species
## 7 Candidiasis
## 8 Candida infection
## 9 Candidiasis, NOS
## 10 CandidosisCondition type == "SYNONYM" is there to remove the redundant fully specified name ‘Candidiasis (disorder)’.
We can also search for those terms in, say, Spanish, by querying the branch containing the Spanish Edition.
concept_descriptions(conceptIds = "78048006", branch = "MAIN/SNOMEDCT-ES/2021-04-30") %>%
.[["78048006"]] %>%
filter(type == "SYNONYM" & lang == "es") %>%
select(term)## term
## 1 infección por especies de Candida
## 2 candidosis
## 3 candidiasis
## 4 moniliasis5. Map SNOMED CT concept codes to ICD-10
SNOMED CT International Edition provides maps to other terminologies and code systems via Map Reference Sets.
Once such map is a map to the World Health Organisation International Classification of Diseases and Related Health Problems 10th Revision (ICD-10). At the time of writing, the Reference Set 447562003 | ICD-10 complex map reference set (foundation metadata concept) | provides a link to zero, one, or several codes of the ICD-10 2016 Edition for concepts descending from:
-
404684003 |clinical finding|, -
272379006 |event|and -
243796009 |situation with explicit context|.
For more information, please consult the ICD-10 Mapping Technical Guide.
For example, let’s extract the ICD-10 code corresponding to 721104000 | Sepsis due to urinary tract infection (disorder) |:
concept_map(map_refset_id = "447562003", concept_ids = "721104000") %>%
select(additionalFields.mapTarget, additionalFields.mapAdvice)## additionalFields.mapTarget
## 1 N39.0
## 2 A41.9
## additionalFields.mapAdvice
## 1 ALWAYS N39.0 | POSSIBLE REQUIREMENT FOR CAUSATIVE AGENT CODE
## 2 ALWAYS A41.9This points us to two ICD-10 codes:
A41.9 Sepsis, unspecified-
N39.0 Urinary tract infection, site not specified, with a disclaimer indicating that the target ICD-10 code can be extended. On examination, the ICD-10 documentation recommends to “use additional code (B95-B98), if desired, to identify infectious agent.”
Conversely, let’s find all SNOMED CT concepts mapped to ICD-10 code N39.0:
concept_map(map_refset_id = "447562003", target_code = "N39.0") %>%
select(referencedComponent.id,
referencedComponent.pt.term,
additionalFields.mapAdvice)## referencedComponent.id
## 1 16916631000119104
## 2 1148925003
## 3 197927001
## 4 721104000
## 5 99631000119101
## referencedComponent.pt.term
## 1 Urinary tract infection caused by Streptococcus agalactiae
## 2 Idiopathic pyuria
## 3 Recurrent urinary tract infection
## 4 Sepsis due to urinary tract infection
## 5 Febrile urinary tract infection
## additionalFields.mapAdvice
## 1 ALWAYS N39.0
## 2 ALWAYS N39.0
## 3 ALWAYS N39.0 | POSSIBLE REQUIREMENT FOR CAUSATIVE AGENT CODE
## 4 ALWAYS N39.0 | POSSIBLE REQUIREMENT FOR CAUSATIVE AGENT CODE
## 5 ALWAYS N39.0 | POSSIBLE REQUIREMENT FOR CAUSATIVE AGENT CODE
## [ reached 'max' / getOption("max.print") -- omitted 23 rows ]We find a total of 28 concepts potentially mappable to N39.0 Urinary tract infection, site not specified.
Note: Complete mapping and semantic alignment with the incoming ICD-11 Mortality and Morbidity Statistics is planned.
References
1 Vasilevsky, Nicole (2018) Introduction to Ontologies. Programming for Biology Workshop, Cold Springs Harbor Laboratory, 26 October 2018, [online] Available from: https://github.com/nicolevasilevsky/CSH_IntroToOntologies/blob/master/IntroToOntologies_CSH_2018-10-28g.pdf (Accessed 29 October 2021)
2 SNOMED International (2021) 2020 Annual Report, [online] Available from: https://www.paperturn-view.com/?pid=MTY165474 (Accessed 15 October 2021)
3 SNOMED International (2021) SNOMED CT: Articulating Stakeholder Value, [online] Available from: https://www.paperturn-view.com/?pid=MTU155774 (Accessed 15 October 2021)
4 NHS Digital (2016) SCCI0034 SNOMED CT Requirements Specifications Amd 35/2016, [online] Available from: https://digital.nhs.uk/data-and-information/information-standards/information-standards-and-data-collections-including-extractions/publications-and-notifications/standards-and-collections/scci0034-snomed-ct (Accessed 15 October 2021)
5 NHS Digital (2021) DAPB4013: Medicine and Allergy/Intolerance Data Transfer Requirements Specification (Amd 5/2021), [online] Available from: https://digital.nhs.uk/data-and-information/information-standards/information-standards-and-data-collections-including-extractions/publications-and-notifications/standards-and-collections/dapb4013-medicine-and-allergy-intolerance-data-transfer (Accessed 15 October 2021)
6 NHS Digital (2021) DAPB4017: Pathology Test and Results Standard Specification (Amd 44/2020), [online] Available from: https://digital.nhs.uk/data-and-information/information-standards/information-standards-and-data-collections-including-extractions/publications-and-notifications/standards-and-collections/dapb4017-pathology-test-and-results-standard (Accessed 15 October 2021)